“This software builds its own hardware”. Craig Venter
Craig Venter made this statement during a TEDxCaltech talk regarding his team’s proof-of-concept research into manufacturing synthetic genomes and having a different host organism ‘boot it up’. The result was a synthetic bacterial genome which, when injected into a host cell, could take control of the cell and have the organism effectively change species.
Syn3.0
A recent research article in Science reports on the most recent developments in this field of synthetic biology. Venter and his colleagues have been working on the organism he discussed in his TEDx talk and have created a minimal genome, an organism with the smallest number of genes that can live and replicate. The premise of the research is reductionist – when we can understand the function of essential genes we can not only replicate viable genomes but develop them from first principles for specific needs.
Syn3.0 was preceded by versions 1 and 2. The article documents the thought and experimental processes that the researchers undertook in developing each and how each development led to the next.
Using Mycobacterium genitalium, being the organism with the smallest known genome, the researchers knocked out genes non-specifically, grew multiple colonies of the resultant mutants and deciphered whether a particular gene was essential (and therefore rarely knocked out in colonies that grew), non-essential (being regularly knocked out in their growing colonies) or quasi-essential (being a gene that was not necessarily required for the resultant organism to be viable but which had adverse effects on its growth and replication times).
In performing this cycle of testing, the genome was segmented into eight pieces. Focusing on each gene in a particular segment, the genes thought non-essential were removed. New genomes containing a mix of the new segments and segments from Syn1.0 (the already reduced genome known to be viable) were created to find which mixture of new and existing segments could make up a viable genome with decent growth rates. The genome made solely of new segments was found non-viable, but a genome with 4 new segments and 4 existing segments with a greater doubling time than Syn1.0 was isolated and studied further.
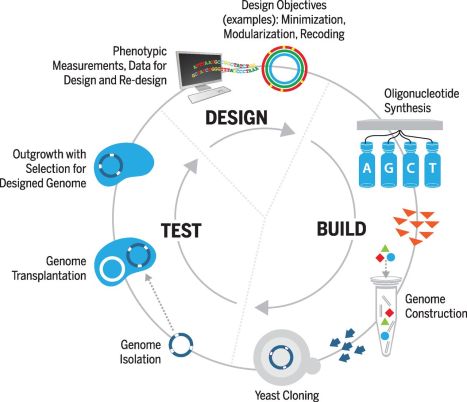
The Design, Build, Test cycle used. Source: AAAS article.
From the starting 525 genes of M. genitalium, they were able to reduce the genome to 375 essential genes and produce a viable organism in Syn3.0.
One issue they had to grapple with was redundant essential genes that were contained within different segments. Where one or the other gene was knocked out without fatal consequences for the cell, the gene was considered non-essential. But this led to the pair of genes being knocked out, leaving the essential gene product missing when the segments were put back together. As a result, the redundant genes had to be identified so that one remained in the genome. This led to the replacement of some genes whilst continuing to find and remove non-essential genes in order to obtain the minimal viable genome that made up Syn3.0.
What we still don’t know
Of the remaining genes, 48% were for the preservation, reading and expression of the genetic information itself, 18% were related to the membrane and its components and 17% to cytosolic metabolism.
149 genes that were found essential to the viability of the cell have an unknown function. These were hypothesised to be for generic proteins whose functions are yet to be deciphered. Searches of gene databases showed some of these to have homologs in other species, but others had no identified homologs within the confines of the searches the researchers performed, showing a notable gap in our knowledge.
Conclusions of the research
By detailing the required genetic functions of a viable cell, the ability to design a cell from scratch and then add functions to it becomes possible. As is suitably detailed within the paper, the environment that the cell grows in will determine what genes are required. In this research the media the cells were grown on were rich in all the essential nutrient sources. For example, as the medium contained suitable amounts of glucose, the genes for glucose transport and metabolism were retained, but genes required to transport and metabolise other forms of carbon weren’t required and could be removed. With this in mind, computational modelling of designed cells should make it easier to quickly design and test novel genomes within particular environments and predict how the cell will behave.
How could this aid agriculture?
Evolution has resulted in some organisms (such as M. genitalium) to be highly adapted to one environment, allowing it to shed genes required to adapt to new environments. Other organisms such as E. coli contain genes enabling them to adapt to different environments should the need arise.
The same can be seen in much of the current research into crop adaptability to drought, salinity stress and pests. Through selective breeding and the use of fertilisers, herbicides, insecticides and the like, our crops have shed genes that would increase adaptability to less than ideal conditions but are no longer conferring any advantage. With the rise of drought problems as an example, researchers are looking to wild, novel and archived cultivars seeking out genes which confer resistance to drought stress that have been lost in our commercial crops.
Although the genome of most crops are significantly larger and more complicated than the M. genitalium worked on in this paper, it does lead to the exciting possibility that we may one day be able to insert a synthetic genome with the required traits into a plant. In doing this, we can adapt it to a particular environment that may not have been amenable to agriculture previously, or to increase yield in environments currently farmed.
It also opens the possibility of creating crops adapted to highly controlled environments. Farming in enclosures that remove the need for genes for resistance to pests or abiotic stress, and using crops stripped of these genes and enhanced for yield in this environemnt, could see increases in food production whilst maintaining or reducing land required for agricultural purposes.
Working on the conclusions of the paper itself, the ability to grow a bacterium containing a gene producing a particular product could in itself be used to produce basic forms of sustenance for use in poverty-stricken areas of the world.
These are ideas that at present are far from reality with many difficult scientific and technological obstacles. But the advances in our basic understanding of what makes a viable genome certainly has some fruit to bear.


This is such exciting science and could really be developed. Well written, and it makes me want to read more into this.
LikeLiked by 1 person